 |
Let us first proceed with the spectral extraction using the source positions found at the IMA step. In our example only the Crab was found by the imaging analysis, so the spectral analysis will generate only the spectrum of the Crab if you run the script with default values (assuming you run j_rebin_rmf in your $REP_BASE_PROD directory).
We are now interested in extracting the spectra in a number of
energy bands different from the ones used for the IMA step in the
previous example. In this case we start a new analysis from COR level with the
desired energy bands for spectral analysis.
If during your analysis the energy bins used
to extract images and spectra are the same, you can continue the
spectral analysis from the same og_group where you previously run
the IMA step, by launching jemx_science_analysis with
startLevel=''SPE'' and endLevel="SPE".
cd $REP_BASE_PROD og_create idxSwg=jmx.lst ogid=crab_spec baseDir="./" instrument=JMX2 cd obs/crab_spec jemx_science_analysis startLevel="COR" endLevel="SPE" \ response="$REP_BASE_PROD/jmx2_rmf_mybins.fits" nChanBins=-4 \ jemxNum=2
The results of the spectral analysis data are in the file
scw/RRRRPPPPSSSF.001/jmx2_srcl_spe.fitsLook on the result of the spectral analysis of the Science Window 010200210010
cd scw/010200210010.001/ fv jmx2_srcl_spe.fitsIn this file, you find the spectra for all sources that were found at the IMA level.

 |
The JEM-X systematics are of the order of a few percents, typically 3%. We add this explicitly to jmx2_srcl_spe.fits file with the command below:
fparkey 0.03 jmx2_srcl_spe.fits SYS_ERR add=yes
The obtained spectrum can be analysed e.g. within XSPEC program:
xspec
XSPEC>cpd /xw
XSPEC>data jmx2_srcl_spe.fits{1}
XSPEC>ign **-3.0 20.0-**
XSPEC>setplot energy
XSPEC>model po
XSPEC>fit
XSPEC>plot ldat del
XSPEC>quit
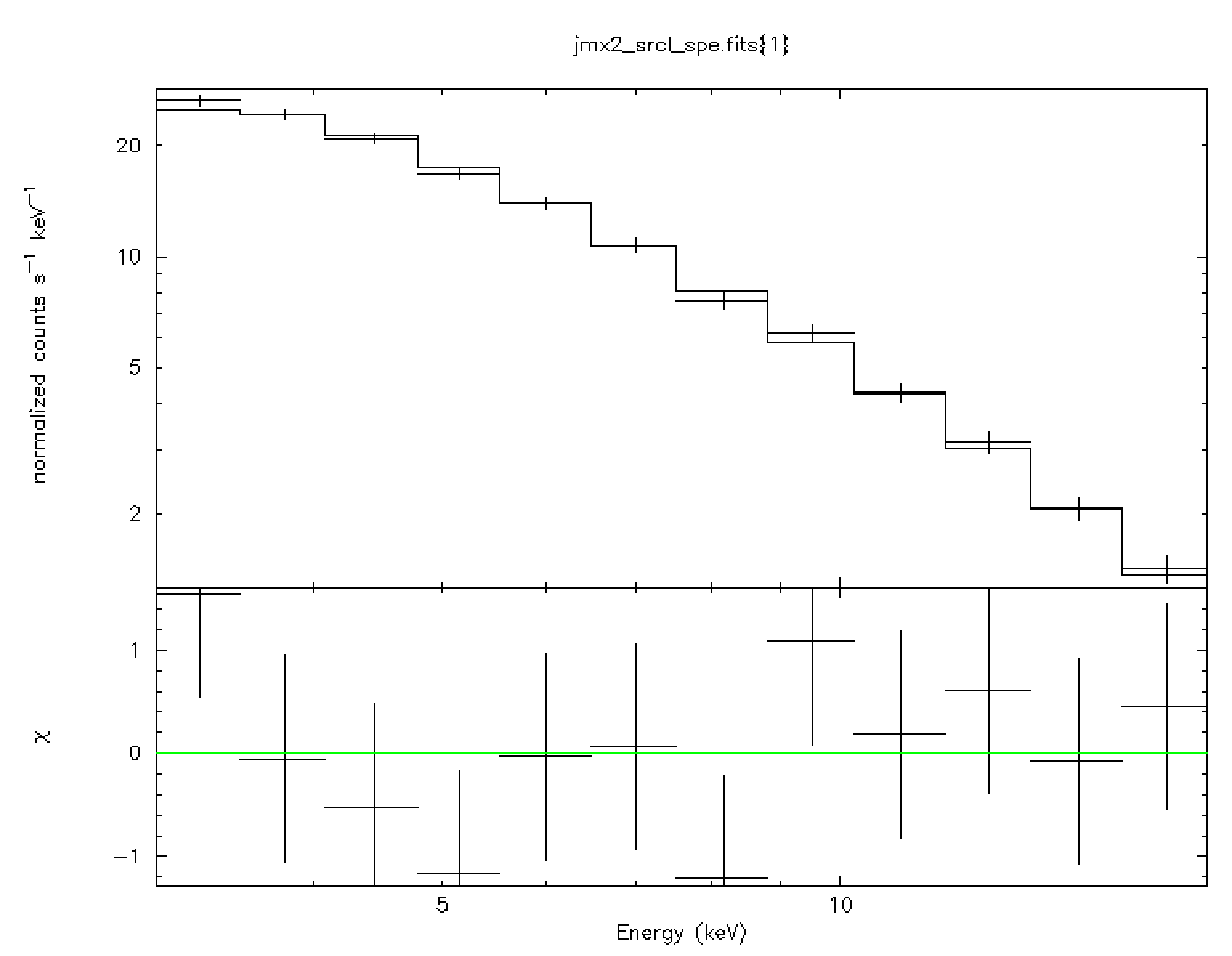
The above set of XSPEC commands reads the data file and fits the data with
``power-law'' model![[*]](footnote.png) . The result is shown in
the left panel of Fig. 19.
. The result is shown in
the left panel of Fig. 19.
To understand the importance of the ``user catalog'', let us extract the Crab spectrum using the catalog position of the source, not the one found at the IMA level. To create the catalog for our source of interest, the Crab, one can just copy the corresponding line from the general reference catalog:
fcopy "${ISDC_REF_CAT}[NAME=='Crab']" user_cat.fits
In the new user_cat.fits file you have to change the FLAG
column to 1; this is the last column of the file, not to be confused with the
JEMX_FLAG for instance. In this case, jemx_science_analysis will
directly copy the source position information to the jmx2_srcl_res.fits
after IMA step (see Sections 6.8.6 and 10.6.1 for an explanation on the possible FLAG values).
Then, you have to re-run the analysis starting from the very beginning, but specifying that you want to use your own catalog for the spectral and lightcurve extraction. Create a new observation group crab_usrcat
cd $REP_BASE_PROD og_create idxSwg=jmx.lst ogid=crab_usrcat baseDir="./" instrument=JMX2copy the user catalog created as explained above into the observation group
cp user_cat.fits obs/crab_usrcatand run the analysis up to the SPE level in this group:
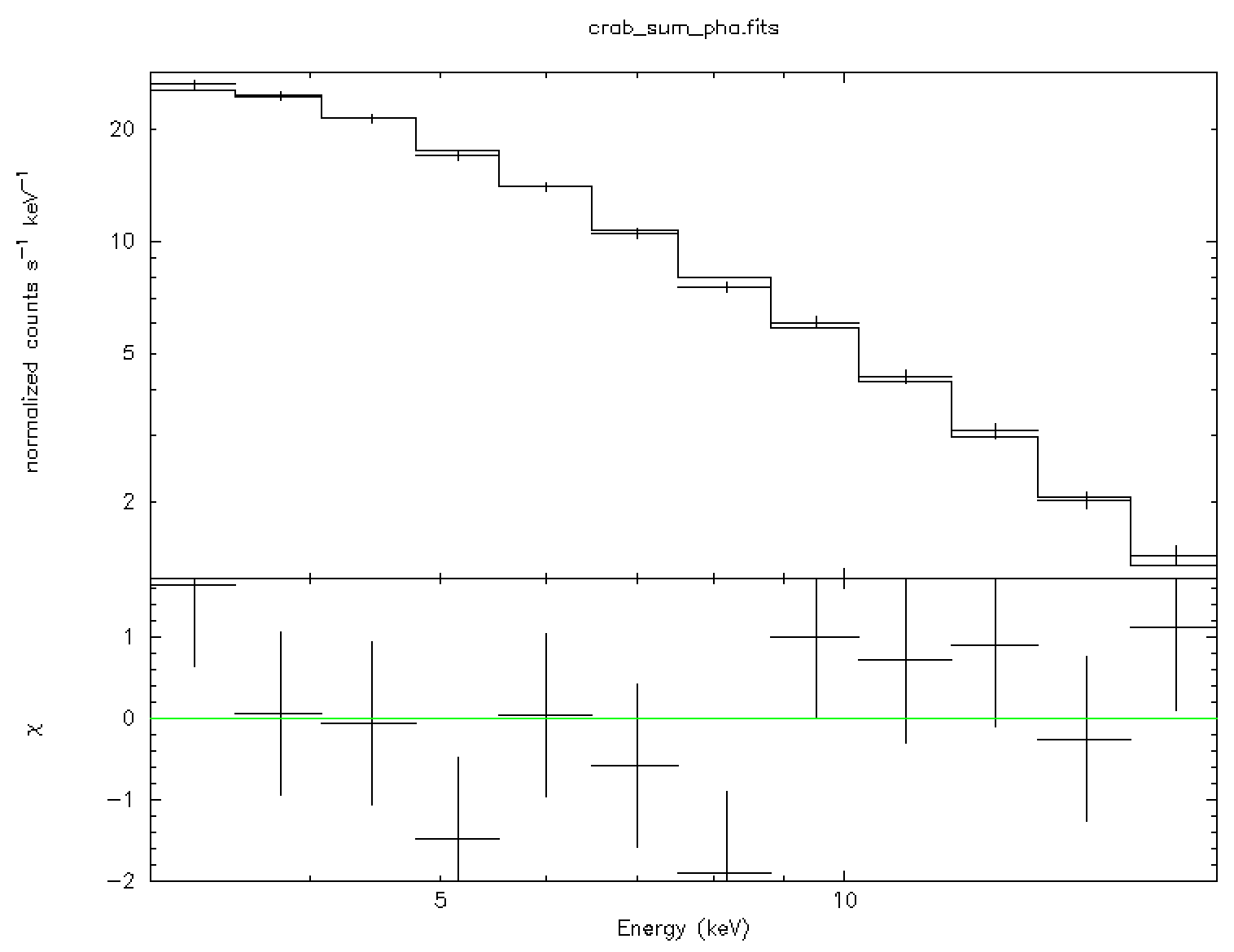
cd obs/crab_usrcat jemx_science_analysis startLevel="COR" \ endLevel="SPE" jemxNum=2 CAT_I_usrCat="user_cat.fits" \ nChanBins=-4 response="$REP_BASE_PROD/jmx2_rmf_mybins.fits"The obtained spectra can be analysed with XSPEC as it was done above. Differences are generally marginal, but they might become more important for a weak source in a crowded region, where the fitted position might be significantly different from the real one.