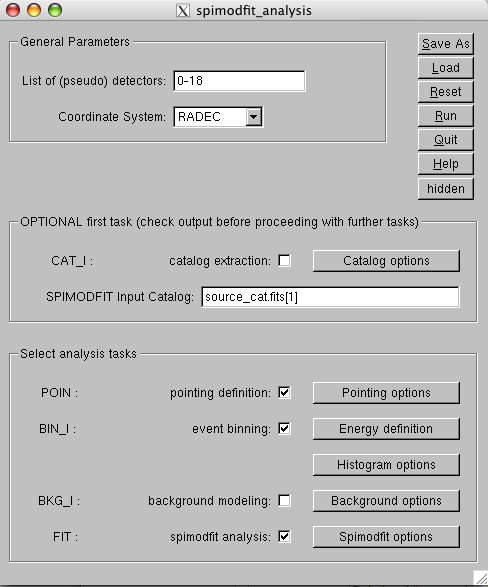
|
Now click on the energy definition options and select the number of channels you want in the final spectrum. We chose 22 logarithmic bins from 25 and 300kev (Fig.![[*]](crossref.png) ) in our example. Close this window, open the window on ``spimodfit options'' (Fig.
) in our example. Close this window, open the window on ``spimodfit options'' (Fig. ![[*]](crossref.png) ) and choose correctly in the upper left frame the selected bins according to your binning choice. You can rebin at this stage with an integer number of bins. All the other parameters can be left to the default values.
) and choose correctly in the upper left frame the selected bins according to your binning choice. You can rebin at this stage with an integer number of bins. All the other parameters can be left to the default values.
Several files are produced as output. One should check the fit quality through, e.g., the log file spi_sa_DATE.log, where DATE is the start time of the analysis. For each energy range, there is the value of the and of the statistics at optimum. These should be around one. If a is obtained, a warning is issued and the user should consider to vary the input catalog and/or the variability time scales.
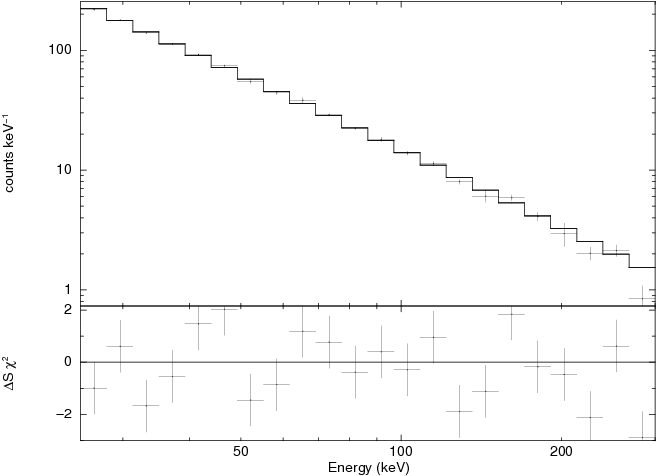
For the user interested in spectral analysis of point sources the relevant output are the spectra named spectrum_SRC.fits and the response matrix spectral_response.rmf.fits. No ancillary effective area is necessary. In our case the Crab spectrum is (spectrum_Crab.fits) whose fit with a power law yields a spectral index
and normalization at 1keV
(Fig. ![[*]](crossref.png) ). The response matrix is computed by the script spimodfit_rmfgen.csh, which should be run manually in case the file spectral_response.rmf.fits is not present.
). The response matrix is computed by the script spimodfit_rmfgen.csh, which should be run manually in case the file spectral_response.rmf.fits is not present.
|
To run the fitting step another time the script automaticcaly deletes the previous output files with the command
rm -f spimodfit\_*
therefore if the user wants to compare the results of subsequent runs, he/she should copy the relevant files elsewhere. At odds with the spi_science_analysis pipeline, the results of spimodfit are not attached to the og_group, therefore the deletion of these files does not produce any harm to the og_tree.