 |
Before starting, you may want to save your imaging analysis again, by copying the current directory
cd .. cp -r spi_analysis spi_imaging_with_catalogue cd spi_analysis
Spiros program can only extract spectra at fixed, known positions. Therefore, a source_cat.fits file must be provided in any case. We assume that you have created/checked a source_cat.fits as explained in last section. Launch ``spi_science_analysis'', un-select the catalogue extraction step, and provide the input catalogue name source_cat.fits in the ``spiros input catalogue'' field.
It is not necessary to run the pointing step again. However, the number of considered energy bins is typically much larger in spectral extractions than in imaging reconstructions. Select the binning, background, and spiros steps. In the energy binning step, you can now select a binning suitable for a spectrum of a bright source with a broader energy range and a relatively high number of bins. For our example, set the number of regions to "1", a "20,1000" keV boundary, and " " as the number of bins. As stated before, using a negative bin number implies a logarithmic binning, with 100 bins from 20keV up to 1MeV in our case.
Open the spiros GUI and set spiros mode to spectra instead of imaging. Click ``Ok'' and ``Run'' the pipeline. After spiros has finished successfully, the pipeline will automatically generate the appropriate response matrix and attach it to your spectra.
To evaluate the obtained results, you can use XSPEC to analyze your spectra. In the following example we fit a single power law from 30 to 1000keV. Here is a verbatim of the XSPEC session.
================================================================================
XSPEC version: 12.8.2
Build Date/Time: Tue Oct 21 17:05:03 2014
XSPEC12>data spectrum_Crab.fits
Spectral Data File: spectrum_Crab.fits Spectrum 1
Net count rate (cts/s) for Spectrum:1 3.912e-01 +/- 3.232e-03
Assigned to Data Group 1 and Plot Group 1
Noticed Channels: 1-100
Telescope: INTEGRAL Instrument: SPI Channel Type: PI
Exposure Time: 1.902e+04 sec
Using fit statistic: chi
Using test statistic: chi
Using Response (RMF) File spectral_response.rmf.fits for Source 1
XSPEC12>cpd /xw
XSPEC12>ign **-30.
11 channels (1-11) ignored in spectrum # 1
XSPEC12>setpl en
XSPEC12>plot lda
XSPEC12>model power
Input parameter value, delta, min, bot, top, and max values for ...
1 0.01( 0.01) -3 -2 9 10
1:powerlaw:PhoIndex>
1 0.01( 0.01) 0 0 1e+20 1e+24
2:powerlaw:norm>
========================================================================
Model powerlaw<1> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 powerlaw PhoIndex 1.00000 +/- 0.0
2 1 powerlaw norm 1.00000 +/- 0.0
________________________________________________________________________
Fit statistic : Chi-Squared = 3.495657e+06 using 89 PHA bins.
XSPEC12>fit 100
<---snip--->
========================================================================
Model powerlaw<1> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 powerlaw PhoIndex 2.11518 +/- 1.13667E-02
2 1 powerlaw norm 11.5598 +/- 0.514568
________________________________________________________________________
Fit statistic : Chi-Squared = 139.58 using 89 PHA bins.
Test statistic : Chi-Squared = 139.58 using 89 PHA bins.
Reduced chi-squared = 1.6044 for 87 degrees of freedom
Null hypothesis probability = 2.988836e-04
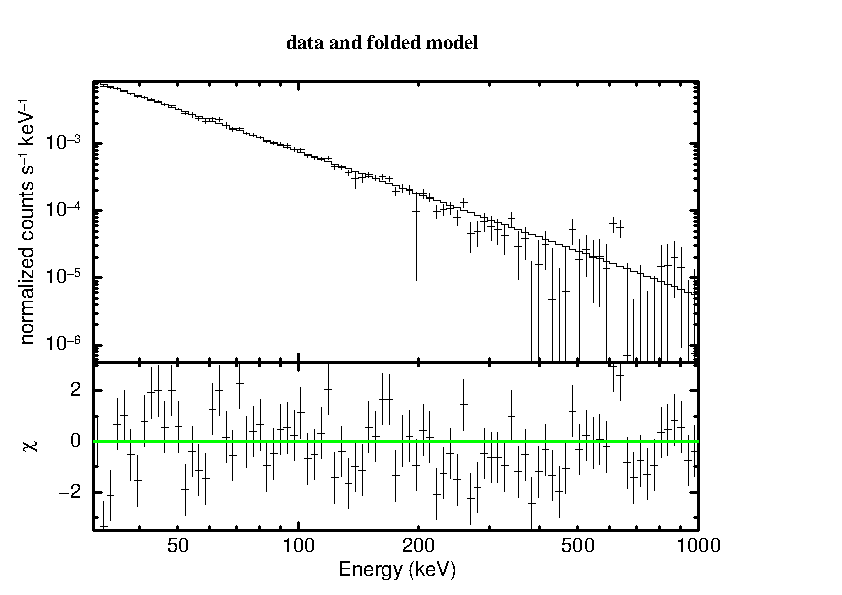
XSPEC12>plot lda del
================================================================================
The final ``plot lda del'' produces the following spectral display:
Note that the remaining deviations are indeed really small, especially considering that we are using only 10 science windows in this example which is a very small number for SPI.
In this section we showed how to analyse SPI data of a relatively
simple case. In fact, Crab is not a highly variable source and in the observations
considered here it was the only bright source in the field of view.
In a typical observation, however, several sources could be located in the field of view of a target source,
and they can be variable. In these cases, the data analysis
requires further settings.
An example is given in Sect. ![[*]](crossref.png) .
.
Section ![[*]](crossref.png) shows how to obtain a reliable spectrum in the energy range 400 keV
2.2 MeV.
shows how to obtain a reliable spectrum in the energy range 400 keV
2.2 MeV.